abdominal trauma detection
rsna kaggle competition
Computed tomography (CT) scans have become crucial for patient evaluation when it comes to injury detection, but interpreting this data can be complex and time-consuming. So, the RSNA Abdominal Trauma Detection Kaggle competition challenged the community to devise a deep learning approach to classify abdominal injuries from multi-phase CT scans in the hopes of assisting medical professionals with diagnosis.
Overview
In this competition, we’re given computed tomography (CT) scans provided by various institutions. The goal is to build a model that can extract critical features within these scans and classify organ injuries (if present) at the liver, spleen, and kidney, as well as any bowel and extravasation injuries.
Pipeline + Model Architecture
In this project, multiple prominent architectures were pipelined together to form several solutions. The major pipelines experimented on within this repository are summarized as follows:
- 2.5D Backbone Feature Extractor → 3D CNN → Prediction Head
- Mask Generator → Merge Input and Mask → 3D CNN → Prediction Head
- Slice Predictor → Input Slice Interpolation → 2.5D Backbone Feature Extractor → 3D CNN → Prediction Head
- Mask Generator → Backbone Feature Extractor (one for input and one for mask) → Merge Input and Mask Features → 3D CNN → Prediction Head
Backbone Feature Extractor
The primary backbone feature extractors utilized were ResNet and Vision Transformer. These architectures are notable for their ability to effectively extract features from visual data through the use of residual connections and self-attention modules (He et al., 2015), (Dosovitskiy et al., 2021). Since the input is a stack of CT scans, it takes the shape \((B, C, H, W)\), where \(B\) is the batch size, \(C\) is the CT scan length, \(H\) is the image height, and \(W\) is the image width. The first thought is to directly apply a 3D CNN, but this would be computationally expensive and memory intensive, especially with high values of \(C\). So, we adopt the 2.5D CNN paradigm depicted below (Avesta et al., 2022), in which we process the CT scans in separate slices and concatenating the extracted features.
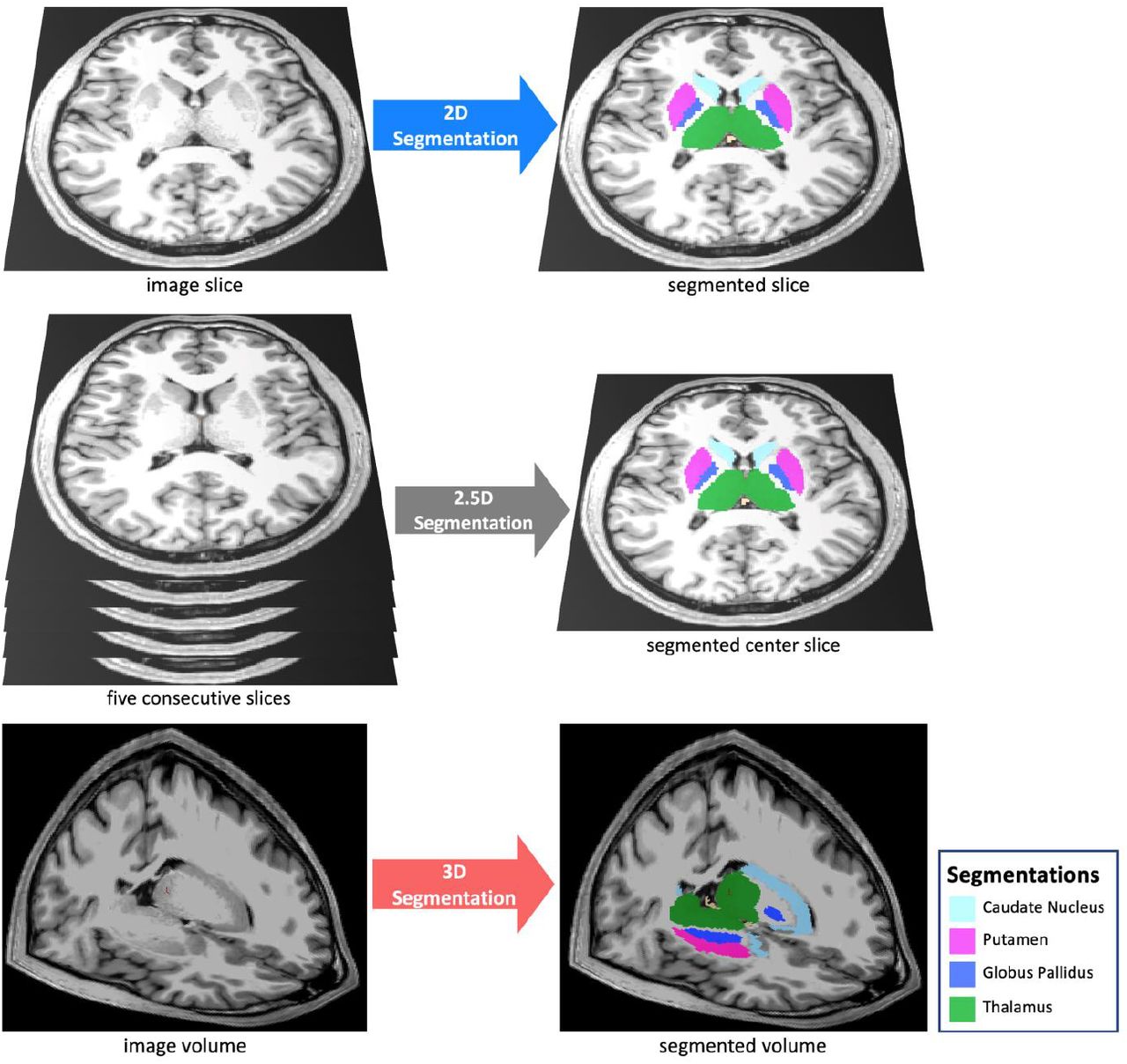
We define a preset slice length \(L\) that represents the number of channels each of these processed slices consist of. Thus, we can process this as follows, where SLICE_CHANNELS = \(L\):
b, c, h, w = scans.shape
x = scans.view(b * (c // SLICE_CHANNELS), SLICE_CHANNELS, h, w)
x = self.backbone(x)
x = x.reshape(b, c // SLICE_CHANNELS, x.shape[-3], x.shape[-2], x.shape[-1])
The backbone can be defined with native PyTorch model definitions. Note that (1) the first convolutional layer must be changed to reflect the chosen slice length and (2) the network head is discarded.
from torchvision.models import resnet18, ResNet18_Weights
backbone = resnet18(weights=None)
self.backbone = nn.Sequential(*(list(backbone.children())[:-2]))
self.backbone[0] = nn.Conv2d(SLICE_CHANNELS, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False)
Mask Generator
The idea for the mask generator component of the pipeline is to predict a mask region for relevant organs to provide the model more context in completing the downstream task of classifying injuries. Both SAM-Med2D and TotalSegmentator were investigated for this mask generation task.
SAM-Med2D
This model is a fine-tuned version on the Segment Anything Model and trained on 4.6 million medical images. SAM-Med2D incorporates domain-specific knowledge from the medical field by adapting adapter layers within the base transformer blocks (Cheng et al., 2023).
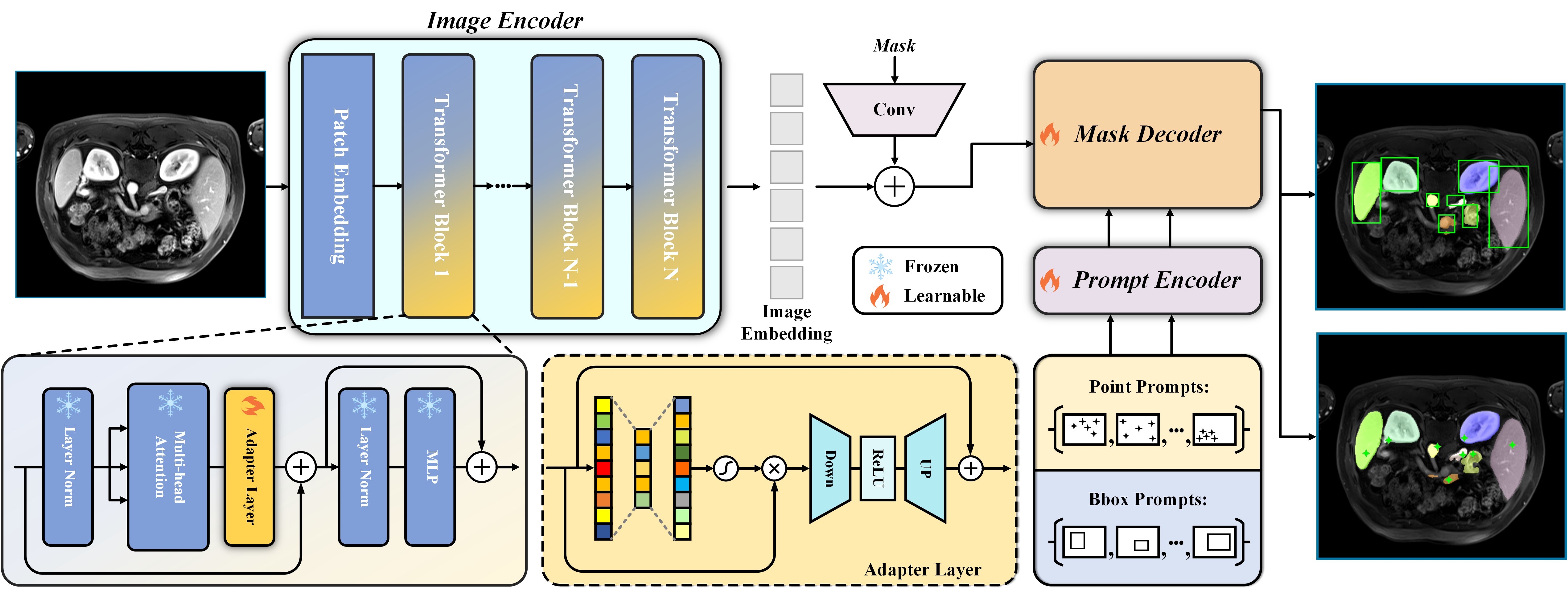
Following the instructions from the official implementation, we can generate and apply the masks as follows:
from SAM_Med2D.segment_anything import sam_model_registry
from SAM_Med2D.segment_anything.automatic_mask_generator import SamAutomaticMaskGenerator
model = sam_model_registry["vit_b"](Namespace(image_size=256, encoder_adapter=True, sam_checkpoint=MASK_MODEL)).to(DEVICE)
mask_generator = SamAutomaticMaskGenerator(model, pred_iou_thresh=0.4, stability_score_thresh=0.5)
def apply_masks(self, id, input):
size = MASK_DEPTH # 12
if id + '.npz' in os.listdir(os.path.join(MASK_FOLDER, self.mode)):
masks = np.load(os.path.join(MASK_FOLDER, self.mode, id + '.npz'))
for i in range(size // 2, N_CHANNELS, size):
input[i - (size // 2):i + (size // 2), :, :] *= masks[str(i)]
else:
save_masks = {}
for i in range(size // 2, N_CHANNELS, size):
image = input[i - 1:i + 2, :, :].transpose(0, 1).transpose(1, 2)
masks = self.mask_generator.generate(image.to(DEVICE))
mask = np.zeros(image.shape[:-1])
for m in masks:
mask = np.where(np.logical_and(m['segmentation'], m['stability_score'] > mask), m['stability_score'], mask)
input[i - (size // 2):i + (size // 2), :, :] *= mask
save_masks[str(i)] = mask
np.savez(os.path.join(MASK_FOLDER, self.mode, id + '.npz'), **save_masks)
In hindsight, I shouldn’t have chosen id and input as variable names. ![]()
The naive approach is to generate the mask and zero out the CT-scan input at non-masked locations. This new masked input can then be feature extracted and concatenated with the features from the original input. This feature vector can then be fused with a multi-layer perceptron. However, another approach would be to implement an attention-based mechanism, which is likely a better utilization of the organ segmentation context. This can be done with a scaled dot product cross attention, where keys \(K\) and values \(V\) are derived from the original input and queries \(Q\) are computed from the segmentation mask.
TotalSegmentator
Because SAM-Med2D falls under the segment anything framework, it is difficult to accurately obtain specific organ segmentations. Instead, we look at TotalSegmentator. This approach is, for the most part, accurate, but suffers from computational inefficiency.
References
2023
- SAM-Med2DJul 2023
2022
2021
- An Image is Worth 16x16 Words: Transformers for Image Recognition at ScaleJul 2021
2015
- Deep Residual Learning for Image RecognitionJul 2015